Anti-Di-Methyl-Histone H3 (Lys9) Antibody, Recombinant Antibody, R2
产品编号:RPTMC5A1-R2
¥ 询价
规格 100ug
产品名称:Anti-Di-Methyl-Histone H3 (Lys9) Antibody, Recombinant Antibody, R2
克隆号:R2
抗体亚型:/
经验证的应用:WB/IP/ChIP/IF/F
交叉反应:Human, Mouse, Rat, Monkey
特异性:Recognizes Di-Methyl-Histone H3 (Lys9)
免疫原:Synthetic peptide
制备方法:/
来源:Recombinant Rabbit IgG
纯化:Protein A purified
缓冲液:Supplied in PBS, 50% glycerol and less than 0.02% sodium azide, PH7.4
偶联物:Unconjugated
状态:Liquid
运输方式:This antibody is shipped as liquid solution at ambient temperature. Upon receipt, store it immediately at the temperature recommended.
储存条件:This antibody can be stored at 2℃-8℃ for one month without detectable loss of activity. Antibody products are stable for twelve months from date of receipt when stored at -20℃ to -80℃. Preservative-Free. Avoid repeated freeze-thaw cycles.
图片:
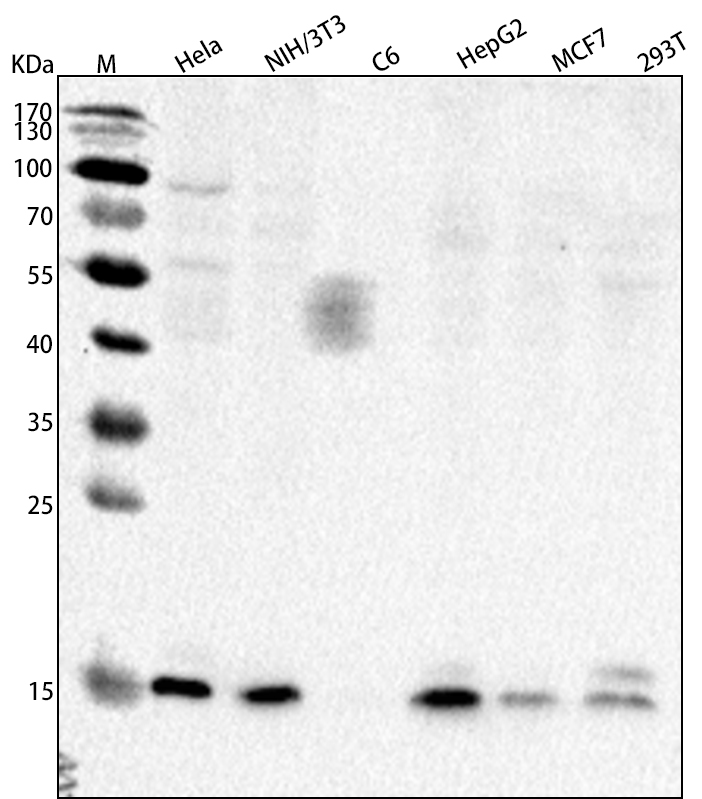
Figure1.Western blot analysisof various cell lines, using RPTMC5A1-R2 at 2ug/ml dilution. Secondary antibody: HRP Goat Anti-Rabbit IgG (H+L) at 1:10000 dilution. Blocking buffer: 5% nonfat dry milk
in TBST. Detection: ECL Basic Kit .
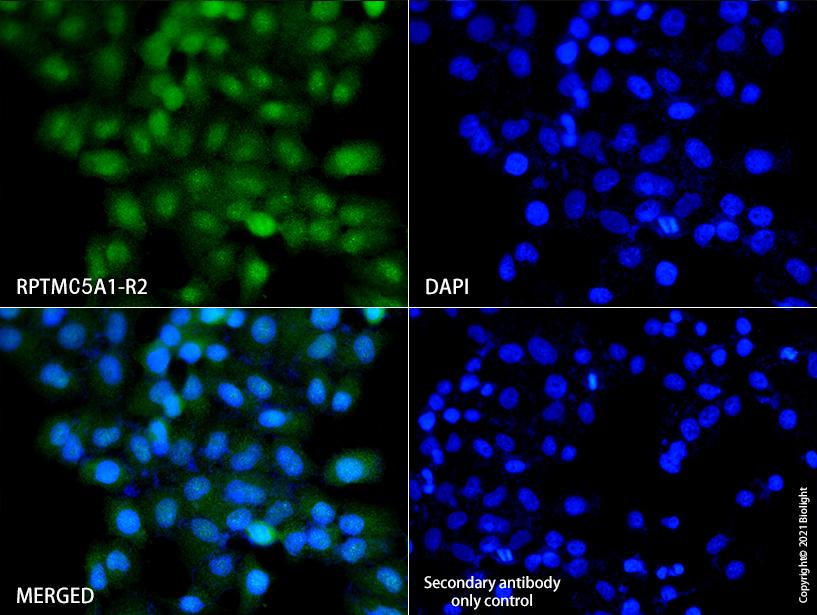
Figure2.Immunofluorescent analysis of Hela cells , using RPTMC5A1-R2 (green) at 10ug/ml dilution. Nuclear DNA was labelled in blue with DAPI (blue).
别称:/
背景信息:The nucleosome, made up of four core histone proteins (H2A, H2B, H3, and H4), is the primary building block of chromatin. Originally thought to function as a static scaffold for DNA packaging, histones have now been shown to be dynamic proteins, undergoing multiple types of posttranslational modifications, including acetylation, phosphorylation, methylation, and ubiquitination. Histone methylation is a major determinant for the formation of active and inactive regions of the genome and is crucial for the proper programming of the genome during development. Arginine methylation of histones H3 (Arg2, 17, 26) and H4 (Arg3) promotes transcriptional activation and is mediated by a family of protein arginine methyltransferases (PRMTs), including the co-activators PRMT1 and CARM1 (PRMT4). In contrast, a more diverse set of histone lysine methyltransferases has been identified, all but one of which contain a conserved catalytic SET domain originally identified in the Drosophila Su(var)3-9, Enhancer of zeste, and Trithorax proteins. Lysine methylation occurs primarily on histones H3 (Lys4, 9, 27, 36, 79) and H4 (Lys20) and has been implicated in both transcriptional activation and silencing . Methylation of these lysine residues coordinates the recruitment of chromatin modifying enzymes containing methyl-lysine binding modules such as chromodomains (HP1, PRC1), PHD fingers (BPTF, ING2), tudor domains (53BP1), and WD-40 domains (WDR5). The discovery of histone demethylases such as PADI4, LSD1, JMJD1, JMJD2, and JHDM1 has shown that methylation is a reversible epigenetic marker.
全称:/
说明书:待上传